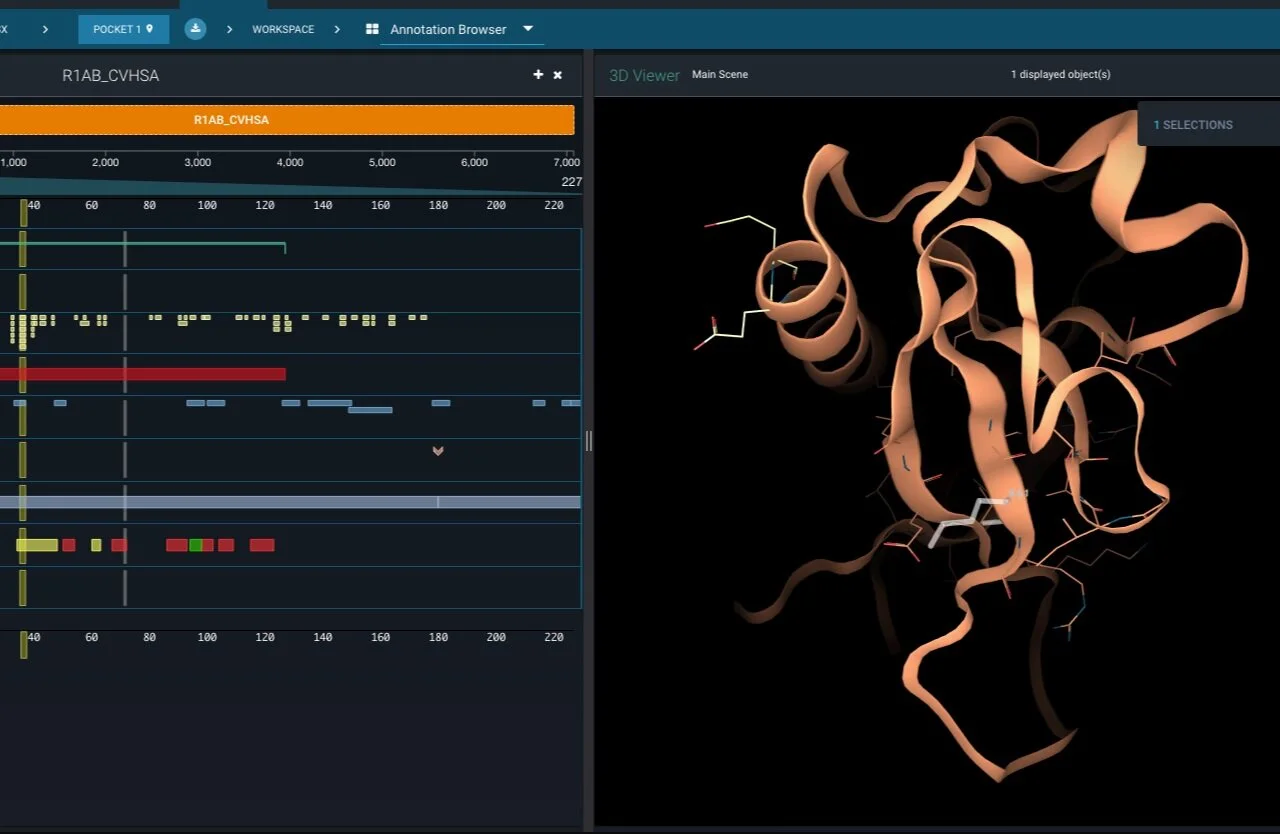
My German cartesian mind (here we go with stereotypes) asks me to start with the first protein expressed by the viral genome of SARS-Cov-2, nsp1. That is, by far, not the low hanging fruit. But it is interesting to learn more about the roles of the different constituents of the virus. Nsp1 actually has also a few very interesting roles to facilitate the viral lifecycle in its host environment.
Read MoreThe bread and butter of every structure based drug design or drug ID effort is the need for 3D structural information of a target of interest related to the phenotypic outcome we want to alter. In the case of SARS-Cov-2 several experimental structures are already available in the RCSB PDB. Other a large scale fragment screens from the Diamond Light Source are slowly becoming available (available in the 3decision protease project for now) on public resources likes the RCSB PDB. Popular homology modelling services also already started to provide several high quality models for some of the yet not resolved structures of the viral proteome.
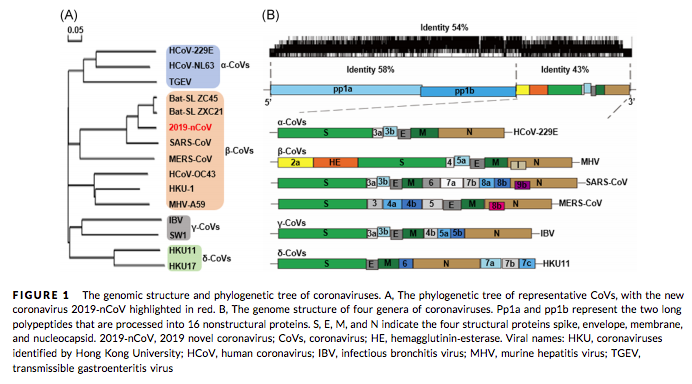
But let’s first have a look what the viral genome actually contains and what are currently pursued efforts.
Read MoreGabriella outlined in previous posts (part I & part II) a few things about SARS-Cov-2. Back at the time the crisis was still far away from our daily lives but things have changed dramatically during the last weeks. After our initial communication to make 3decision.discngine.cloud freely available for all Covid-19 related collaboration projects we were contacted independently by several volunteers from academic and industrial groups to collaborate on a global effort. At the mean time a lot of novel initiatives have seen the day and everybody is publishing blog posts (like I am right now), linkedIn articles, chemarxiv or bioarxiv preprints etc. It’s a little bit a scientific wild-west happening right now on a global scale.
Read MoreThis time around you won’t see any source code. Sorry about that, next time maybe.
Here I’ll talk more about sometimes overlooked aspects when developing applications for R&D in life sciences. So I guess this post might be useful to our competitors or people developing applications within large corporations in the life science & pharma industry … anyway ;)
I’ll take the example of our current 3decision developments. You might have heard or read about 3decision already before. If not, check it out - 3decision .. it’s super cool!
Read MoreI love MOE, it’s a great piece of software, but unfortunately it’s not free and now that I’m nor in academia, nor in a pharma company I find myself unable to further develop an approach I implemented in the past which is kind of frustrating to put it politely. The same goes for Amber.
So I started to look for alternatives, also under a gentle demand from Anthony (poking me regularly) who wanted to apply the whole approach in his fragment screening pipeline. That’s in the scope of the crazy impressive XChem project. If you haven’t heard / read about that check it out - it gives you a bit of an insight on what will be possible very near future in xray crystallography!
I think that’s my first RDKit post! So reason to celebrate!
Here' I’ll focus on a very nice feature available in Open Source Software and very useful in daily structure based but even ligand based drug design tasks.
The problem
How can I dock small molecules into a receptor and avoid as much as possible the “what is the right pose” problem?
Read MoreA brief walkthrough on how to compile the molfile plugin for VMD in order to use it for programming and integrating into your own code.
Read MoreIf you ever had to manage, administer or install a Pipeline Pilot server you know that this is usually not a piece of cake. It's a fairly long process, involves a lot of more or less manual steps (that you could automate, but did you do it so far?)
Read MoreWe decided to reiterate our support to the German Conference on Chemoinformatics to be held in Fulda in November this year.
Read MoreNetwork Collection 2016 brings Formal Concept Analysis to your desktops!
Read MoreChemistry Collection 2016 is out now!
Read MoreDiscngine Sponsor of the German Conference on Chemoinformatics in Fulda!
Read MoreDiscngine Chemistry Collection 2.0 for Pipeline Pilot is available now!
Read MoreWith the very fresh 2.0 release of the Discngine Chemistry Collection for Pipeline Pilot nifty new functionality is available to Pipeline Pilot users. Find out more about the new release of the Chemistry Collection.
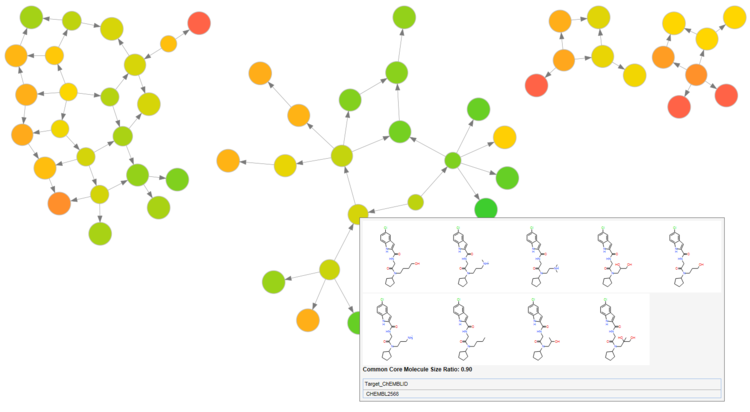
This post describes one of these new features, fuzzy context specific matched molecular pairs (fcsMMPs), why you should adopt them and a few examples of what you can do with this. Note that this post focuses on target activity prediction (fairly unsual for MMP analysis), but everything that is described here applies to more classical property prediction too.
Everything that is shown here can be done with this Pipeline Pilot Component Collection. Even the data is included.
If you do not feel 100% familiar with the concepts behind matched molecular pairs analysis, please read through the introductory part below. If you know what a transformation, a common core and a context is and have done matched pairs effect analysis in the past, you can directly go to the fcsMMP specific part of this post.
Read MoreF1000Prime Associate Faculty Membership : Good or Bad ?
Read MorePBS is widely used as queuing system on small size to huge clusters. This is just a little post to resume somehow all installation steps necessary to get a single node torque server running on Centos 6.4 (64bit).
Read MoreDiscngine sponsor of Accelrys Accelerate 2014 in Washington, D.C.!
Read MoreHappy New Year 2014!
Read MorePeter and Vincent from Discngine attended this year German Conference on Chemoinformatics.They presented a poster there about our recent developments on Matched Molecular Pairs!
Read More